
Learning programming games in Java is a project-based learning approach that addresses real-world development challenges. Besides providing hands-on practice, coding games in Java polish problem-solving skills and boost creativity. Java offers a fertile ground for budding developers. It allows them to create, innovate, and learn.
Many platforms, like DoMyCoding, offer comprehensive courses for learning the language. But if you wish to learn it independently, we have a solution for that, too. This article delves into how one can leverage the games coded in Java to polish their skills.
Understanding Java’s Strength for Game Development
Java gaming program is a popular choice among users who have no prior experience in coding. Some of the key reasons for this popularity are listed below.
1. Platform Independence
Java is a versatile programming language, that allows a single code to run anywhere. A Java game code is compatible with all platforms, including Windows, MacOS, Linux, or a web browser.
2. Object-oriented Programming
A Java game code is easier to manage and maintain. This benefit is only facilitated by its unique object-oriented programming.
3. Diverse Libraries
Java boasts a variety of frameworks that are suitable for all developers. For instance, the Lightweight Java Game Library can be used for coding games for beginners who have no prior experience.
Getting Started with Java Game Development
Before moving ahead with game development, it is important to understand and set up the development environment. Install Java, and choose an integrated development environment (IDE). This provides tools for code editing, debugging, and testing. Let’s begin with the steps of Java game development.
Install Java
Java Installation is a prerequisite to learning simple Java game coding. Follow the steps mentioned below to install Java.
- Download Java for your operating system (after checking minimum requirements)
- Run the installer
- Set the environment path variable to point to the installation directory
- Verify the installation
Guess the number game explanation
Let’s start game development using Java programming, as in Figure 1. The first two lines import two Java classes, Random and Scanner, for random number generation and taking user input, respectively.
Line # 4 defines the main Java class ‘Guessing_game’ for the number guessing game. Line # 6 defines a private Random Java class object.
Line # 8 defines the main Java method from where code execution starts. Line # 10 defines the Scanner Java classes. Lines # 12-16 define some variables and set parameters for this game, e.g., the game generates random numbers between 1 and 50 only.
Line # 21 initiates a do-while loop that prompts the user to guess the number. Note that the integer variable random_number at line # 14 has already stored the result of random number generation. scanner.nextInt() function at line # 23 reads the user input.
The conditional block if-else if-else at lines # 26-34 compares the user input with the random number and provides feedback to the user, simultaneously incrementing the variable tries that keeps a record of the number of attempts made by a user.
The last line of the Java game code, line # 35, defines the condition for loop termination; the do-while loop terminates only when a user has made the right guess.

Java class and fields
Since Java is an object-oriented programming language, class is its fundamental component. Java class is a container that encapsulates data and Java methods.
Every Java class has its unique data variables and Java methods. The data encapsulated by the program is formally called the Java field. Preceded by an access modifier (public, protected, or private), the Java field characterizes the class object. A private field is accessible only to the Java methods of that class.
Running and packaging the game
A Java package corresponds to a structured hierarchy of directories to save the source code, Java class, and a manifest file. An IDE automatically makes a Java package, but for understanding, let’s go through individual steps assuming a Linux environment.
Step1. Create a project folder and make a sub-folder to store the source code file (game.java). Following are the commands to do this.
$ mkdir src
$ mv sample.java src/game.java
Step 2. Create a directory tree following the package name.
$ head -n1 src/game.java
package com.example_1.game;
$ mkdir -p com/example_1/game
Step 3. Create a manifest.txt file.
echo "manifest-Version: 1.0" > manifest.txt
Step 4. Compiling the game into a Java class produces Main.class in com/example_1/game
$ javac src/game.java -d com/example_1/game
$ ls com/example_1/game/
Main.class
Step 5. The jar command packages the application into a Java Archive (JAR).
$ jar cfme game.jar \
manifest.txt \
com.example_1.game.Main \
com/example_1/game/Main.class
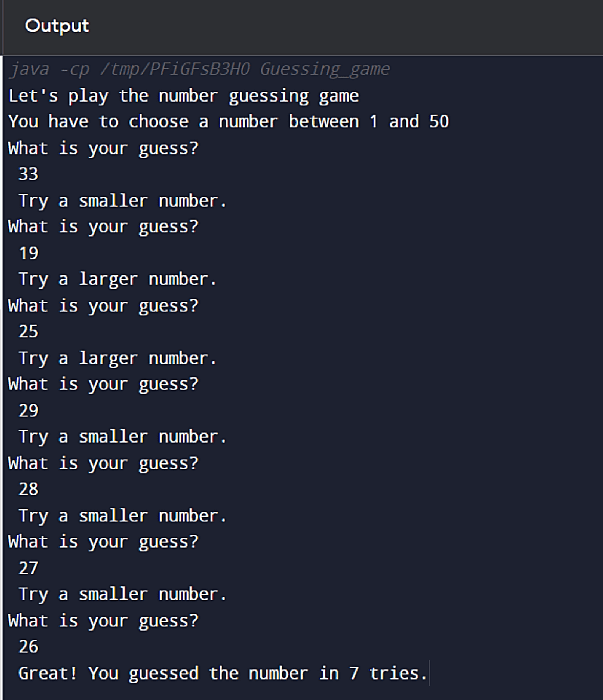
Step 6. Finally, run the Java programming file. Figure 2 shows the output.
$ java -jar game.jar
Package declaration and import statements
Java package declaration is a good practice as it assigns a unique identifier to each Java game project.
Import statements simplify Java programming using classes from other namespaces and provide access control.
The first two lines of code in Figure 1 simply import two classes, Random and Scanner, from the package java.util. This improves code readability, whereas the other approach of defining the complete class Random makes the code unnecessarily long.
Pseudo-random numbers and main method
Pseudo-random numbers in Java programming are unlike the true random numbers, i.e., they can be deterministic. Random number generation requires a random seed to generate a random (unpredictable) number. See line # 6 of Figure 1, where the Java class Random is instantiated to generate the seed.
There must be at least one main Java class in a long Java game code because this is the program’s entry point. There is only one class in this case, so it is the main class with the main method. The main method must be public and static.
Application logic and syntax
Basics of Java syntax include Java classes, Java methods, variables, and control structures. Remember the use of semi-colon after every statement. Basic logic involves conditional statements and loops used in the game’s code.
The guessing game in Figure 1 simply declares a main class and defines one Java field and Java method. The application asks the user to input a number, which it compares with Java’s own random number generator, and it continues until the user inputs the correct value. Java syntax is almost the same as in C++. System.out.println() is a print statement used to keep track of the program’s flow.
What are the Core Game Development Concepts?
Beginning with Java, coding games can be a lot easier with a clear understanding of the basic concepts. Similar to the concept of arrays in Java, developers should be familiar with the gaming concepts. The key concepts related to Java programming games are listed below.
1. Game Loop
Every game has a game loop, which controls the game state updates and render cycles. It guarantees that the game proceeds by changing element placements, checking for user inputs, and rendering the modified state on screen.
2. Rendering graphics
JavaFX includes the Canvas and GraphicsContext classes for drawing graphics. Learning how to use these for game element rendering, animation handling, and scene management is important.
3. Handling User Input
User input, whether by keyboard or mouse, determines game interactions. JavaFX's event-handling methods enable developers to capture and respond to user inputs, thereby managing game elements accordingly.
4. Game Physics
While basic games programmed in Java may not require advanced physics, understanding collision detection and response is critical. This entails identifying when game elements interact (for example, a player character picking up an object) and processing these interactions properly.
Conclusion
Learning Java through game development, such as creating a "Guess the Number" game, offers a hands-on approach to learning programming skills, including problem-solving and creativity. This method involves installing Java, understanding its basic concepts such as classes, methods, loops, and conditional statements, and executing a project from start to finish.
Through this hands-on project, you can learn the importance of planning, the intricacies of game mechanics, and the power of Java's libraries and tools in bringing creative ideas to life. The journey of creating a game in Java is a stepping stone to a fulfilling career in technology, where the only limit is your imagination and willingness to learn.
Frequently Asked Questions
-
Can I code a game in Java?
Yes, Java is a general-purpose programming language for developers. Games in Java can easily be coded using simple programming codes.
-
Is Java better than C++?
-
Is Java good for 2D games?
-
Is Java coding free?
















